Global epistasis emerges from a generic model of a complex trait (or: random walks on a hypercube with reweighting)
I still don’t really know what global epistasis is, but I really enjoyed Gautam Reddy‘s talk about this paper at the CMSA Big Data conference two weeks ago. Here’s a simple model of evolution. There are n genes (x_1, … x_n) which can each be in one of two conditions, which we label as 1 and -1; so there are now 2^n genotypes which we have modeled as the vertices of a hypercube. You have some population of organisms and you start them off at some vertex. And then mutation happens at a constant rate, which means each organism sets off on a random walk.
But that’s not evolution yet. We need some notion of fitness, which we model as a function f on the hypercube, given by some polynomial
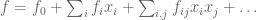
And organisms with higher fitness reproduce more; so you reweight at each time step, placing more probability on those walkers whose current vertex has higher fitness. And then you keep track of the mean reproduction rate as the walk progresses. It turns out that you typically get sharp improvement followed by diminishing returns, and that the shape of the curve is a good match for what’s observed in actual experiments with actual organisms. (Did you know there’s an ongoing experiment in E. coli evolution that started in 1988 and has now tracked over 80,000 generations of bacteria? Amazing.)
What’s more, there is lots of interesting math that goes into understanding what we can learn about the function f (which can’t be directly observed) from the changes in reproduction rate (which can.) In particular — though this is the part I’m not sure I totally followed — I think they were saying that experimental results aren’t really compatible with f having no cross-terms; in other words, one can see that there have to be a lot of terms like x_1 x_2, where the sign of the effect of changing x_1 depends on the value of x_2, and vice versa. I think this is what “epistasis” means.
Reddy’s co-author Michael Desai (who has his own myriad-generational population of yeast bubbling away in his lab) is teaching a really interesting multi-disciplinary double-counted undergraduate course for Harvard students interested in the modern life sciences.
The course uses examples from biology as an integrating theme, principles from physics and mathematics to reduce complex problems to simpler forms, and computer simulation to allow students to develop their intuition about the behavior of the dynamical systems that control the physical and biological universe. The course includes bootcamps to introduce students to biological experiments and the computer language, Python. Each semester will include a project lab, in which students will work in small teams to do original research on unsolved biological problems.
Sounds fun!
Jordan Ellenberg's Blog
- Jordan Ellenberg's profile
- 411 followers



