Science for Authors: How DNA Identification Works #scienceforauthors

So, I'm going to kick off one of these science related posts that I hope you all find helpful and easy to understand. If you have any ideas for future posts, and of course any questions, please shout it out in the comments!
When you watch the TV shows that link a suspect with a crime scene sample based on DNA, what are they actually doing? What are these markers that they're talking about? Well, that can be a bit tricky, depending on the scenario, but for today we're going to talk about the most common system: the CoDIS (Combined DNA Index System) markers.
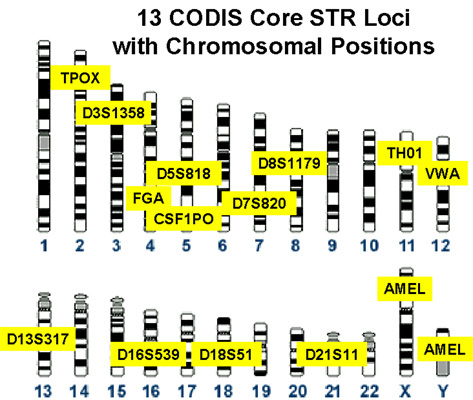
To start this off, we have to talk a little about what STRs are (aka Short Tandem Repeats, sometimes knowns as microsatelites). These are locations in everyone's genome where little pieces of DNA repeat over and over. A huge portion of our genome (as much as 50%!) is made up of bit of repeating DNA that doesn't necessarily code for anything (unlike a gene, which codes for a protein--but keep in mind that this doesn't mean this is useless DNA, even thought it used to be referred to as "junk" DNA). Anyhow, these little pieces of DNA (which are several base-pairs, or the basic element that makes up DNA: A, T, C, or G) repeat over and over, with most people having a different set of repeats. It looks a bit like this:
ATCG ATCG ATCG ATCG ATCG
So that's five repeats of the pattern ATCG. Now, remember that humans have two copies of each of their chromosomes (46 total, or 23 pair). So, we would look at one of these locations in the genome and count how many repeats are found on each of the chromosomes. So, a person would have a pattern that might look like this:
ATCG ATCG ATCG ATCG ATCG ATCG ATCG
ATCG ATCG ATCG
In this case, their repeat motif would be 7, 3 at this position in the genome. For the markers that are used in law enforcement in the USA (which differ a little bit for different countries around the world), we look at 13 different places around the genome. So, a genetic ID would have readings for each of these different locations that look like this:
7,3
4,5
12,14
22,15
13,13 (where both chromosomes have the same # of repeats, or are homozygous)
The chances of having the 7,3 repeat motif might be something like 1 out of every million people have that pattern. The chances of having the 4,5 repeat motif at another spot in the genome may be around 1 in 100,000. The chances of having both are actually (1/1,000,000)(1/100,000)--or the chances of both multiplied together. So the chances of having the exact repeating motif for all the 13 markers gets pretty dang small, to where it's probably only a single person on the planet with that exact same number of repeats at each of those spots.
Unless you're an identical twin :)
These readings are what you're looking at if someone flashes something like this on the screen, trying to look official:
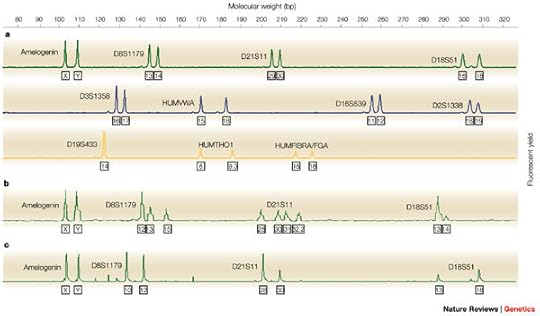 Each of those peaks tell a reader how many copies of the repeat motif there are at each location in the 23 pair of chromosomes.
Each of those peaks tell a reader how many copies of the repeat motif there are at each location in the 23 pair of chromosomes.So, how does one make a match? Well, if a suspect's DNA was found at a crime scene, we'd expect there to be the same repeating motif at each of the markers for BOTH samples: the one we took from the suspect and the one from the scene.
But what if the repeats don't match? Well, that means that the DNA from the suspect is not a match to the one from the crime scene. Not that this means they didn't do it, but the sample could be from someone else.
What if only some of the repeats match? If half of them do, we'd expect the sample to come from a full sibling or parent of the person (this is how paternity tests work). A more distant relative may also share some matches. But, this can get tricky, and the statistical power of saying who the sample came from gets much more complex :)
Okay, so this is a basic run-down of things. I hope it makes some sense though! Shout out any questions in the comments!
Published on April 25, 2016 04:00
No comments have been added yet.



