Michael J. Behe's Blog, page 29
October 31, 2022
At SciTech Daily: Traces of Ancient Ocean Discovered on Mars – This Means a “Higher Potential for Life”
Penn State University researchers write:
New evidence for an ancient northern ocean on Mars has been uncovered in a recently released set of topography maps. These maps offer the strongest case yet that the planet once experienced sea-level rise consistent with an extended warm and wet climate, which was far different than the harsh, frozen landscape that exists today.
 Stitched together from 28 images, this view from NASA’s Curiosity Mars rover was captured after the rover ascended the steep slope of a geologic feature called “Greenheugh Pediment.” In the distance at the top of the image is the floor of Gale Crater, which is near a region called Aeolis Dorsa that researchers believe was once a massive ocean. Credit: NASA/JPL-Caltech/MSSS.
Stitched together from 28 images, this view from NASA’s Curiosity Mars rover was captured after the rover ascended the steep slope of a geologic feature called “Greenheugh Pediment.” In the distance at the top of the image is the floor of Gale Crater, which is near a region called Aeolis Dorsa that researchers believe was once a massive ocean. Credit: NASA/JPL-Caltech/MSSS.“What immediately comes to mind as one the most significant points here is that the existence of an ocean of this size means a higher potential for life,” said Benjamin Cardenas, assistant professor of geosciences at Penn State and lead author on the study recently published in the Journal of Geophysical Research: Planets. “It also tells us about the ancient climate and its evolution. Based on these findings, we know there had to have been a period when it was warm enough and the atmosphere was thick enough to support this much liquid water at one time.”
Whether Mars had an ocean in its low-elevation northern hemisphere has long been debated in the scientific community, Cardenas explained. Using topography data, the research team was able to show definitive evidence of a roughly 3.5-billion-year-old shoreline with substantial sedimentary accumulation, at least 900 meters (3,000 feet) thick, that covered hundreds of thousands of square kilometers.
Using software developed by the United States Geological Survey, the research team generated the maps with data from the National Aeronautics and Space Administration (NASA) and the Mars Orbiter Laser Altimeter. They discovered over 6,500 kilometers (4,000 miles) of fluvial ridges and grouped them into 20 systems to show that the ridges are likely eroded river deltas or submarine-channel belts, the remnants of an ancient Martian shoreline.
On Earth, the ancient sedimentary basins contain the stratigraphic records of evolving climate and life, explained Cardenas. If researchers want to locate a record of life on Mars, an ocean as big as the one that once covered Aeolis Dorsa would be the most logical place to start.
“A major goal for the Mars Curiosity rover missions is to look for signs of life,” Cardenas said. “It’s always been looking for water, for traces of habitable life. This is the biggest one yet. It’s a giant body of water, fed by sediments coming from the highlands, presumably carrying nutrients. If there were tides on ancient Mars, they would have been here, gently bringing in and out water. This is exactly the type of place where ancient Martian life could have evolved.”
The stratiographic analysis these researchers carried out to show records of large waterways on Mars is certainly interesting and valuable science. But, to borrow a phrase, what immediately comes to mind as one of the most significant points here, is that to assume that the existence of surface water on a planet can somehow naturally lead to life is to depart from the realm of established science.
Copyright © 2022 Uncommon Descent . This Feed is for personal non-commercial use only. If you are not reading this material in your news aggregator, the site you are looking at is guilty of copyright infringement UNLESS EXPLICIT PERMISSION OTHERWISE HAS BEEN GIVEN. Please contact legal@uncommondescent.com so we can take legal action immediately.Plugin by Taragana
October 29, 2022
At Quanta Magazine: How Gödel’s Proof Works
His incompleteness theorems destroyed the search for a mathematical theory of everything. Nearly a century later, we’re still coming to grips with the consequences.
Natalie Wolchover writes:
In 1931, the Austrian logician Kurt Gödel pulled off arguably one of the most stunning intellectual achievements in history.
Mathematicians of the era sought a solid foundation for mathematics: a set of basic mathematical facts, or axioms, that was both consistent — never leading to contradictions — and complete, serving as the building blocks of all mathematical truths.
But Gödel’s shocking incompleteness theorems, published when he was just 25, crushed that dream. He proved that any set of axioms you could posit as a possible foundation for math will inevitably be incomplete; there will always be true facts about numbers that cannot be proved by those axioms. He also showed that no candidate set of axioms can ever prove its own consistency.
His incompleteness theorems meant there can be no mathematical theory of everything, no unification of what’s provable and what’s true. What mathematicians can prove depends on their starting assumptions, not on any fundamental ground truth from which all answers spring.
Undecidable questions have even arisen in physics, suggesting that Gödelian incompleteness afflicts not just math, but — in some ill-understood way — reality.
The article next outlines a “simplified, informal rundown of how Gödel proved his theorems.” What the results imply is discussed next.
No Proof of ConsistencyWe’ve learned that if a set of axioms is consistent, then it is incomplete. That’s Gödel’s first incompleteness theorem. The second — that no set of axioms can prove its own consistency — easily follows.
What would it mean if a set of axioms could prove it will never yield a contradiction? It would mean that there exists a sequence of formulas built from these axioms that proves the formula that means, metamathematically, “This set of axioms is consistent.” By the first theorem, this set of axioms would then necessarily be incomplete. But “The set of axioms is incomplete” is the same as saying, “There is a true formula that cannot be proved.”
Gödel’s proof killed the search for a consistent, complete mathematical system. The meaning of incompleteness “has not been fully fathomed,” Nagel and Newman wrote in 1958. It remains true today.
“Truth is bigger than proof.”
Copyright © 2022 Uncommon Descent . This Feed is for personal non-commercial use only. If you are not reading this material in your news aggregator, the site you are looking at is guilty of copyright infringement UNLESS EXPLICIT PERMISSION OTHERWISE HAS BEEN GIVEN. Please contact legal@uncommondescent.com so we can take legal action immediately.Plugin by Taragana
Review: J. W. Thornton (2022): Simple mechanisms for the evolution of protein complexity
Review: J. W. Thornton (2022): Simple mechanisms for the evolution of protein complexity
https://reasonandscience.catsboard.com/t2706-main-topics-on-proteins-and-protein-synthesis#9622
Proteins are tiny models of biological complexity: specific interactions among their many amino acids cause proteins to fold into elaborate structures, assemble with other proteins into higher-order complexes, and change their functions and structures upon binding other molecules.
Comment: Merriam-Webster describes the word “elaborate” as: “planned or carried out with great care, to produce by labor,
Synonyms: Adjective: complex, complicated, detailed, fancy, intricate, involved, sophisticated
Its hard to overlook the teleological aspect of the word. We can all agree that proteins are well-described as elaborate: Nucleopores for example have been described recently as “a massive complex of roughly 1,000 proteins that helps channel DNA instructions to the rest of the cell” 12
These complex features are classically thought to evolve ( so they are not certain) via long and gradual trajectories driven by persistent natural selection. But a growing body of evidence from biochemistry, protein engineering, and molecular evolution shows that naturally occurring proteins often exist at or near the genetic edge of multimerization (MULTIPROTEIN COMPLEXES), allostery, and even new folds, so just one or a few mutations can trigger acquisition of these properties. These sudden transitions can occur because many of the physical properties that underlie these features are present in simpler proteins as fortuitous by-products of their architecture. Moreover, complex features of proteins can be encoded by huge arrays of sequences, so they are accessible from many different starting points via many possible paths. Because the bridges to these features are both short and numerous, random chance can join selection as a key factor in explaining the evolution of molecular complexity.
Comment: What Thornton et.al. conveniently do not mention, is that the origin of protein complexity does not start when life starts, but there has to be already a fully developed proteome to kickstart life. The question of how it all started from a random bunch of almost infinite different disordered molecules laying around prebiotically is a much harder task.
R. Mukhopadhyay (2013): Researchers have a better grasp of the processes of selection and evolution once a function appears in a peptide. “Once you have identified an enzyme that has some weak, promiscuous activity for your target reaction, it’s fairly clear that, if you have mutations at random, you can select and improve this activity by several orders of magnitude,” says Dan Tawfik at the Weizmann Institute in Israel. “What we lack is a hypothesis for the earlier stages, where you don’t have this spectrum of enzymatic activities, active sites and folds from which selection can identify starting points. Evolution has this catch-22: Nothing evolves unless it already exists.3
A minimal amount of instructional complex information is required for a gene to produce useful proteins. Minimal size of a protein is necessary for it to be functional. Thus, before a region of DNA contains the requisite information to make useful proteins, natural selection would not select for a positive trait and play no role in guiding its evolution.
Understanding how living things acquired their complex features—structures and functions that arise from specific interactions among differentiated parts—has been a central aim of biology for centuries. 1
Comment: Thornton directs to the first paper with link no.1. Interesting, what in the introduction is being confessed: ” The consensus among evolutionists seems to be (and has been for at least a century) that the morphological complexity of organisms increases in evolution, although almost no empirical evidence for such a trend exists. ( Thank you for pointing that out!) Most studies of complexity have been theoretical, and the few empirical studies have not, with the exception of certain recent ones, been especially rigorous; reviews are presented of both the theoretical and empirical literature. The paucity of evidence raises the question of what sustains the consensus, and a number of suggestions are offered, including the possibility that certain cultural and/or perceptual biases are at work. ( I could not agree more)
Darwin supplanted divine agency with the evolutionary view: complexity arises through “numerous successive, slight modifications” under the influence of natural selection, because each step enhances functions that contribute to fitness. This scenario of gradual elaboration and optimization is well-supported in numerous cases.4-7
Comment: Thornton now links to a few science papers, that supposedly back up the claim that ” gradual elaboration and optimization is well-supported in numerous cases “. Shall we have a look if that is indeed factual ?
K. E. Jones (2019): A fundamental concept in evolutionary biology is that life tends to become more complex through geologic time, but empirical examples of this phenomenon are controversial.
Comment: Something, between being well-supported, and being controversial, is quite a stretch if you ask me.
One debate is whether increasing complexity is the result of random variations if there are evolutionary processes that actively drive its acquisition, and if these processes act uniformly across clades.
Comment: We know that there is pre-programmed adaptation, but that does not say anything in regard to the transition from one phenotype to a completely different one with complex novelties, like limbs, eyes etc. ( which is what in the end is in dispute ) For more, see here: Non random mutations : How life changes itself: the Read-Write (RW) genome
The mammalian vertebral column provides an opportunity to test these hypotheses because it is composed of serially-repeating vertebrae for which complexity can be readily measured. Here we test seven competing hypotheses for the evolution of vertebral complexity by incorporating fossil data from the mammal stem lineage into evolutionary models. Based on these data, we reject Brownian motion (a random walk) and uniform increasing trends in favor of stepwise shifts for explaining increasing complexity. We hypothesize that increased aerobic capacity in non-mammalian cynodonts may have provided impetus for increasing vertebral complexity in mammals.
Comment: hypothesize and may have are not words used to describe confirmed and demonstrated facts, but characterizes speculation and guesswork!
At the very end of the paper, in the section “Discussion”, the paper ends with: “Selection for higher activity levels combined with the release of respiratory constraints in cynodonts may have provided the trigger required to achieve vertebral complexity, and the subsequent biomechanical and ecological diversification of the presacral column in mammals”
Comment: If you were expecting more than speculation, you will be disappointed. If one wants to explain the origin of vertebra, the mechanisms involved in vertebra development have to be elucidated first,. Anatomical comparisons are entirely meaningless.
To understand novelty in evolution, we need to understand organisms down to their individual building blocks, down to the workings of their deepest components, for these are what undergo change.
Dr. Marc W. Kirschner: The Plausibility of Life: Resolving Darwin’s Dilemma 2005
Osteogenesis is in fact an irreducibly complex process, and therefore, evolution is inadequate to explain its origin, as elucidated here: Origin and development of bones ( Osteogenesis)
Furthermore, in order to explain the origin of such complex structures as a vertebra, which requires billions of cells, one has to explain each step, as i do here:
How do biological multicellular complexity and a spatially organized body plan emerge?
The most famous is the modern vertebrate eye, which evolved from a simple light-sensitive precursor by sequentially adding cell types and more complicated relationships among tissues, each of which improved visual sensitivity or acuity.
Comment: Before addressing the evolution of vertebrate eyes, Thornton would do good to explain as well how eyespots (which are the starting point of Nilsson’s paper) originated. I have yet to see a plausible account for that. Evolutionnews published my article on that, see here: The Evolution of the Eye, Demystified and then explain the origin of the visual cycle: Origin of phototransduction, the visual cycle, photoreceptors and retina and the human eye: How the origin of the human eye is best explained through intelligent design
In the last half-century or so, a pageant of intricate forms has been revealed at a tiny new scale. Every protein is itself a complex system, because its physical and functional features depend on a large number of interactions among its many constituent amino acids. For example, a protein’s ability to fold into its native tertiary structure depends on complementary steric, electrostatic, and hydrophobic interactions among scores or hundreds of residues.
Comment: Orchestrating these forces that permit the right folding that in the end will result in proteins with specific functions requires foresight and – intelligence: Forces Stabilizing Proteins – essential for their correct folding
The same is true of quaternary structure: most proteins assemble with other molecules into specific multimeric complexes, and the interfaces that hold these complexes together often involve dozens of tightly packed residues with a high degree of electrostatic and steric complementarity. Another form of complexity is allostery—changes in a protein’s function caused by binding an effector molecule—which typically involves many amino acids to bind the effector and coupleing of binding to the active site.
Comment: instantiating “Molecular Recognition Through Structural Complementarity” requires as well intelligence, as i point out here
During the last ~3.8 billion years, evolution has generated proteins with thousands of different folds, unique multimeric interactions, and varying modes of allosteric regulation.
Comment: Orchestration and timing of cellular processes require life-essential, precise timing, cross-regulation, coordination, the right sequence of processes, the right speed, at the right rate, there are checkpoint mechanisms, error checking, and repair at various stages. Question: What emerged first: protein synthesis, or the right, precise coordination of the whole process, and its respective proteins and signaling processes doing the job? For more, see here
This diversity presents a molecular version of the classic question about the evolution of biological complexity: how did the stepwise processes of evolution repeatedly produce complicated systems from simpler precursors? Darwin’s model of gradual adaptive elaboration was developed to explain morphological and physiological complexity, but it has been assumed to apply as well to the evolution of complex molecular features. Intuitively, the view that protein complexity always evolves by long, consistently adaptive trajectories may seem sensible or even necessary, given certain assumptions. For a feature to evolve, the sequence states that encode it must arise by mutation and then be fixed in populations.
Comment: Must be? Yes. If we presuppose philosophical naturalism and do not permit design in the explanatory framework as a possible alternative explanation.
Multimerization, allostery, and protein folds all involve elaborate arrays of interacting amino acids, so how else could they have been acquired if not by a long and specific series of many sequence changes? And, in turn, how could we explain the fixation of a long series of particular mutations if each step were not driven by the deterministic power of selection?
Comment: Does natural selection have deterministic power? Does it determine something? No, it does not. Natural selection is not an acting force but is passive. It does not invent something new.
R.Carter:‘Natural selection’ properly defined simply means ‘differential reproduction’, meaning some organisms leave more progeny than others based on the mutations they carry and the environment in which they live. 56
It has been suggested that such features might arise neutrally, but fixation by chance alone would be vanishingly improbable if many particular mutations are required. Recent advances in protein biochemistry and molecular evolution call into question the assumptions that underlie the argument for the gradual adaptive evolution of protein complexity. Of particular note are dramatic improvements in protein design, deep mutational scanning (which characterizes the functions of huge numbers of protein sequence variants), and ancestral protein reconstruction (which uses phylogenetics to infer the sequences of ancient proteins and experiments to determine the molecular functions and structures that existed in the deep past). This new body of work shows that just one or a few mutations can drive the acquisition of multimerization, allostery, and even new folds from natural precursors that lack these features; furthermore. It also explains why these short paths exist: simpler proteins often already possess most of the physical properties that underly these features. Moreover, the networks of sequences that yield multimerization, allostery, or a given protein fold appear to be immense, and they are closely intercalated at numerous places with the sequence networks of functional proteins that lack the feature. As a result, proteins can—and do—acquire new complex features by neutral processes. Contrary to the metaphor underlying the gradualist view, the complex features of proteins are not singular, massive mountain peaks that an evolving protein can climb only via a long trek under the deterministic engine of natural selection. Rather, many complex features are better conceived of as innumerable wrinkles, each small enough to be mounted in a single step (or just a few), which proteins repeatedly encounter as they wander through a vast multidimensional landscape of functional amino acid sequences.
Comment: Even if they do, so what? Does that explain biocomplexity and diversification, and the origin of organismal and organ and limb novelties?
(1) massive/infinite amounts of “natural” small changes;
(2) many beneficial/magical, random genetic mutations, producing novel, more complex structures over eons of time, from a “simple” virus or amoeba, to man; and
(3) Natural Selection – the continued survival (of fittest) of these different/new living Species (cf. speciation) & Families. ?
Macroevolution. Fact, or fantasy?
Another point: Imagine a production line in a factory. Many robots there are lined up, and raw materials are fed into the production line. The materials arrive at Robot one. It processes the first step. Then, when ready, the product moves on and is handed over to the next Robot. Next processing step. And that procedure repeats 17 times. In the end, there is a fully formed subpart, like the door of a car. That door is part of a larger object, like the finished car. That door by its own has no use unless mounted at the right place in the car. Nobody would project a car door without visualizing the higher end up front, in the project and development stage, and based on the requirement, specify the complex shape of the door which precisely will fit the whole of the chassis of the car where it will be mounted. And the whole production line and each robot the right placement and sequence where each robot will be placed must be planned and implemented as well. Everything has to be projected with a higher-end goal in mind. And there is interdependence. If one of the robots ceases to work for some reason, the whole fabrication ceases, and the completion of the finished car cannot be accomplished. That means, a tiny mal connection of one of the robots in the production line of the door might stop the production of the door, and the finished car cannot be produced.
Metabolic pathways in cells are analogous to human-made production lines. They work in an integrated fashion together. If one enzyme or protein mutates to produce a different product, than that new product might not be useful for the next manufacturing step, and the whole thing breaks down.
Alone, and individually, each of the 17 enzymes that synthesize Chlorophyll can do nothing. But together, lined up like in a factory production line, they process the intermediate substrates, hand it over to the next, and the next, 17 manufacturing steps, one enzyme machine fine-tuned to produce the substrate, which the next enzyme can process, that in the end, makes the chlorophyll molecule, the most abundant organic compound found on earth, in bacteria, plants, algae, diatoms, plankton, corals and, oxygenating the oceans, make abundant marine life possible.
Chlorophyll by its own can do nothing. But together with many other Chlorophylls in the antenna complex, it can transfer, when energized to a higher energy state through photons, its resulting high-energy electron state to its adjacent Chlorophyll, and so down to the reaction center, and energize the P680 special Chlorophyll pair, and start the electron chain. But the lineup and order of these Chlorophylls cannot be just so. It must be just right. Each Chlorophyll must have the right distance, one from the other, to produce the energy transfer. And what an energy transfer that is – it’s an engineering marvel !! It’s almost 100% efficient, and done by quantum mechanical Förster resonance energy transfer principles !!! The stupendous ingeniosity cannot be enough outlined…. Human-made solar panels, in comparison, have just 20% efficiency…
The reaction center IMHO cannot operate and release an electron, if it does not find its replacement – which the oxygen-evolving complex provides.
If the oxygen-evolving complex would be not there, no oxygen would be released into the atmosphere and no respiration could occur, and no advanced life exist !!
Chlorophyll by its own in the antenna complex, will produce triplet states and burn the membrane where they are embedded. But Carotenoid chromophores do join them, and prevent triplet states to occur – they quench the solar energy when too strong, and release it as heath.
Yup, No Carotenoids, and you would probably not be here to read my lines. To make Carotenoids, it is another extremely complex biosynthesis process, but that’s another story….
On the one side, you have an intelligent agency-based system of irreducible complexity of tight integrated, information-rich functional systems which have ready on-hand energy directed for such, that routinely generate the sort of phenomenon being observed. And on the other side imagine a golfer, who has played a golf ball through a 12-hole course. Can you imagine that the ball could also play itself around the course in his absence? Of course, we could not discard, that natural forces, like wind, tornadoes, or rains or storms could produce the same result, given enough time. the chances against it, however, are so immense, that the suggestion implies that the non-living world had an innate desire to get through the 12-hole course.
1. Shelly Fan: In Its Greatest Biology Feat Yet, AI Unlocks the Complex Proteins Guarding Our DNA June 14, 2022
2. Nuclear pore complexes. Design, or evolution ?
3. Rajendrani Mukhopadhyay: “Close to a miracle” Sept. 23, 2013
4. Katrina E. Jones: Stepwise shifts underlie evolutionary trends in morphological complexity of the mammalian vertebral column 07 November 2019
Plugin by Taragana
Extreme genome repair, and remarkable morphogenesis by self-assembly point to design
Extreme genome repair, and remarkable morphogenesis by self-assembly point to design
https://reasonandscience.catsboard.com/t2061p275-my-articles#9626
Extreme Genome Repair (2009): If its naming had followed, rather than preceded, molecular analyses of its DNA, the extremophile bacterium Deinococcus radiodurans might have been called Lazarus. After shattering of its 3.2 Mb genome into 20–30 kb pieces by desiccation or a high dose of ionizing radiation, D. radioduransmiraculously reassembles its genome such that only 3 hr later fully reconstituted nonrearranged chromosomes are present, and the cells carry on, alive as normal 1
T. Devitt (2014): John R. Battista, a professor of biological sciences at Louisiana State University, showed that E. coli could evolve to resist ionizing radiation by exposing cultures of the bacterium to the highly radioactive isotope cobalt-60. “We blasted the cultures until 99 percent of the bacteria were dead. Then we’d grow up the survivors and blast them again. We did that twenty times,” explains Cox. The result were E. coli capable of enduring as much as four orders of magnitude more ionizing radiation, making them similar to Deinococcus radiodurans, a desert-dwelling bacterium found in the 1950s to be remarkably resistant to radiation. That bacterium is capable of surviving more than one thousand times the radiation dose that would kill a human. 2
Simple bacteria can restart their ‘outboard motor’ by hotwiring their own genes (2015):
Unable to move and facing starvation, the bacteria evolve a replacement flagellum – a rotating tail-like structure that acts like an outboard motor – by patching together a new genetic switch with borrowed parts. When an organism suffers a life-threatening mutation, it can rapidly rewire its genes. The remarkable speed with which old genes take on new tasks suggests that life has unexpected levels of genetic flexibility. In theory, the bacteria should have starved to death and effectively gone extinct. Yet over the course of a weekend, they managed to patch themselves back together with borrowed genes.” Scientists made the discovery by accident while researching ways to use naturally occurring bacteria to improve the yield of crops. A microbe was engineered so that it could not make its ‘propeller-like’ flagellum and forage for food. However, when a researcher accidentally left the immotile strain out on a lab bench, the team discovered the bacteria had evolved over just a few days. The new variety of bacteria had resurrected their flagella in the process.
Remarkably, this happened because the mutants had rewired a cellular switch, which normally controls nitrogen levels in the cell, to activate the flagellum. This rescued these bacteria, which faced certain death if they didn’t move to new food sources. The bacteria being studied, Pseudomonas fluorescens, are among a group of bacteria scientists are researching for use in agriculture, as a kind of ‘plant probiotic’. These could help crops grow or fight off diseases, leading to higher yields. However, a key problem is that the bacteria lack resilience, as their positive effects can stop working after only a short period of time. Dr Jackson, a microbiologist at Reading, said: “Plant probiotics could make crops grow more reliably in the future, helping to feed the world’s growing population. This new study shows that these bacteria are more resilient than previously thought, as they show a remarkable capacity to overcome catastrophic changes and find a way to survive. “This gives us crucial insights into how bacteria could survive and change, and the challenge now is to see if this occurs in their natural soil and plant environment.” 3
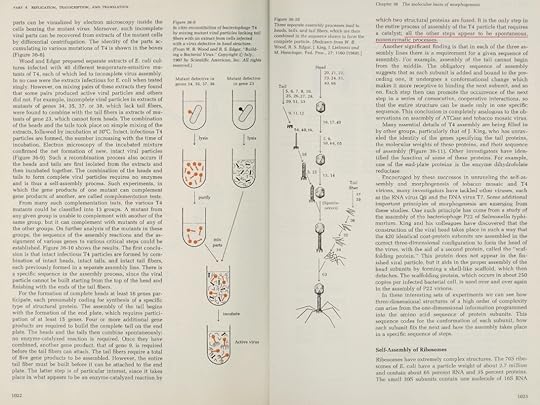
K. Eric Drexler: Engines of Creation 2.0 ( 2006): The T4 phage, acts like a spring-loaded syringe and looks like something out of an industrial parts catalog. It can stick to a bacterium, punch a hole, and inject viral DNA (yes, even bacteria suffer infections). Like a conqueror seizing factories to build more tanks, this DNA then directs the cell’s machines to build more viral DNA and syringes. Like all organisms, these viruses exist because they are fairly stable and are good at getting copies of themselves made. Whether in cells or not, nanomachines obey the universal laws of nature. Ordinary chemical bonds hold their atoms together, and ordinary chemical reactions (guided by other nanomachines) assemble them. Protein molecules can even join to form machines without special help, driven only by thermal agitation and chemical forces. By mixing viral proteins (and the DNA they serve) in a test tube, molecular biologists have assembled working T4 viruses. The machinery of the T4 phage, for example, self-assembles from solution, apparently aided by a single enzyme.…self-assembling structures (…For a description of molecular self-assembly, including that of the T4 phage and the ribosome, see Chapter 36 of Lehninger’s Biochemistry)
This ability is surprising: imagine putting automotive parts in a large box, shaking it, and finding an assembled car when you look inside! Yet the T4 virus is but one of many self-assembling structures. (M. YANAGIDA, 1984: The virus particle contains more than 3,000 protein subunits of some 30 polypeptide species !!) Molecular biologists have taken the machinery of the ribosome apart into over fifty separate protein and RNA molecules, and then combined them in test tubes to form working ribosomes again. To see how this happens, imagine different T4 protein chains floating around in water. Each kind folds up to form a lump with distinctive bumps and hollows, covered by distinctive patterns of oiliness, wetness, and electric charge. Picture them wandering and tumbling, jostled by the thermal vibrations of the surrounding water molecules. From time to time two bounce together, then bounce apart. Sometimes, though, two bounce together and fit, bumps in hollows, with sticky patches matching; they then pull together and stick. In this way protein adds to protein to make sections of the virus, and sections assemble to form the whole. 4
E. V. Koonin, the logic of chance, page 376: Breaking the evolution of the translation system into incremental steps, each associated with a biologically plausible selective advantage is extremely difficult even within a speculative scheme let alone experimentally. Speaking of ribosomes, they are so well structured that when broken down into their component parts by chemical catalysts (into long molecular fragments and more than fifty different proteins) they reform into a functioning ribosome as soon as the divisive chemical forces have been removed, independent of any enzymes or assembly machinery – and carry on working. Design some machinery that behaves like this and I personally will build a temple to your name! 5
AlphaFold (2020) What a protein does largely depends on its unique 3D structure. Figuring out what shapes proteins fold into is known as the “protein folding problem”, and has stood as a grand challenge in biology for the past 50 years. In a major scientific advance, the latest version of our AI system AlphaFold has been recognized as a solution to this grand challenge by the organizers of the biennial Critical Assessment of protein Structure Prediction (CASP). 6
Comment: Imagine the engineering effort that it would take for protein engineers to produce nanomachines that would need no nano arms and nano hands to assemble complex nanomachines, but design parts that would be able to assemble on their own just by shaking them, like motors, bearings, and moving parts coming together randomly, and then self-assemble into a fully operational nano-machine. The engineers would need to know the single individual forces and how they would interact with the forces from the other parts. The problem becomes even more apparent when we consider that one of the forces that influence proteins is for example Van der Waals forces which operate based on quantum mechanical principles. R. W. Newberry (2019): The dominant contributors to protein folding include the hydrophobic effect and conventional hydrogen bonding, along with Coulombic interactions and van der Waals interactions. Human technology and advance is far from being able to design this. What a feat would THAT be!
1. Rodrigo S. Galhardo: Extreme Genome Repair 2012 Apr 4.
2. T. Devitt: In the lab, scientists coax E. coli to resist radiation damage March 17, 2014
3. WEEKEND EVOLUTION: BACTERIA ‘HOTWIRE THEIR GENES’ TO FIX A FAULTY MOTOR 26 February 2015
4. K. Eric Drexler: Engines of Creation 2.0 ( 2006)
5. E. V. Koonin, The logic of chance (2012), page 376
6. AlphaFold: a solution to a 50-year-old grand challenge in biology November 30, 2020
 Copyright © 2022
Uncommon Descent
. This Feed is for personal non-commercial use only. If you are not reading this material in your news aggregator, the site you are looking at is guilty of copyright infringement UNLESS EXPLICIT PERMISSION OTHERWISE HAS BEEN GIVEN. Please contact legal@uncommondescent.com so we can take legal action immediately.
Copyright © 2022
Uncommon Descent
. This Feed is for personal non-commercial use only. If you are not reading this material in your news aggregator, the site you are looking at is guilty of copyright infringement UNLESS EXPLICIT PERMISSION OTHERWISE HAS BEEN GIVEN. Please contact legal@uncommondescent.com so we can take legal action immediately.Plugin by Taragana
October 28, 2022
At Reasons.org: Bear Species Hybridization Shows God’s Providence
Fazale Rana writes:
Adaptability—the capacity to manage change is an invaluable trait in today’s ever-changing work environment. Adaptable workers are resilient, curious, and resourceful. They are willing to experiment and to risk failure. Most importantly, they understand the big picture, always keeping it at the forefront of everything they do. Some people are innately adaptable; others aren’t. Still, those who aren’t adaptable by nature can develop the qualities that help them to thrive on the job.
Adaptability is also a valuable quality in biology. In fact, many biologists believe that adaptability is one of the universally descriptive features of life. Organisms are exquisitely suited for their environments. Yet the environment changes. And like adaptable employees who can navigate workplace changes, organisms have the means to adapt to a shifting landscape. Those organisms that respond to change will persist; those that can’t will disappear.
Biologists have discovered a variety of mechanisms that operate at a population level that enable species to adapt to: (1) changes in the environment, (2) predatory pressure, and (3) fluctuating resources. The mechanism list includes: (1) natural selection, (2) sexual selection, and (3) genetic drift.
 Image: Reasons.org
Image: Reasons.orgRecently, a large team of collaborators, headed by researchers from the University of California, Santa Cruz, highlighted another mechanism that they think contributes to organisms’ ability to adapt: introgression—the introduction of genetic material into the gene pool of another species through interbreeding or hybridization.1 Insights like this one are often viewed as prima facia evidence for life’s evolutionary history. But this discovery can also be viewed legitimately from a creation model standpoint, where adaptability reflects God’s providential care for his creation. In other words, God has designed the world so that populations of organisms have the innate capacity to adapt and ensure that they will survive and thrive.
Polar Bear and Brown Bear Introgression
More evidence for the connection between introgression and adaptability became available when the UC Santa Cruz-led team examined the genome of a polar bear fossil specimen that age-dates between 70 and 110 thousand years ago. This specimen consists of a jawbone recovered from the beach near the Beaufort Sea by Port McLeod in Arctic Alaska.
From the ancient DNA extracted from one of the polar bear’s fossilized teeth, the team reconstructed high-quality sequences for the nuclear and mitochondrial genomes. The sequence data indicates that this specimen was indeed a polar bear but its genetic fingerprint falls outside the genetic diversity range for extant polar bears.
Comparison of this ancient polar bear genome with the genomes of extant brown bears indicates that the population to which the polar bear belonged interbred with a group of brown bears, around 100,000 years ago. As it turns out, these ancient recipients of the polar bear genetic material became ancestral to all brown bears living today. In fact, about 10% of the contemporary brown bear genome comes from this ancient introgression.
Introgression and Adaptation
One of Charles Darwin’s most important scientific accomplishments was to identify a mechanism to account for the origin of species. In the process, he demonstrated that species aren’t fixed entities but can change through natural and sexual selection.
These two mechanisms, along with genetic drift, allow populations of organisms to adapt. This adaptation can occur in one of two ways: (1) through changes in standing genetic variation in the population with the frequency of the alleles in the population changing in the response to environmental changes, or (2) through mutations that introduce new alleles altogether. The former mechanism leads to rapid response to environmental changes; the latter mechanism requires much more time to effect change.
Based on the recent work by the UC Santa Cruz-led investigators (along with other studies), introgression can be added to the list of mechanisms that serve as drivers for adaptive change.2 Adaptive introgression can rapidly introduce a large amount of new genetic information into a population across multiple genetic loci. The result is a response to environmental changes that’s more rapid than mutations afford and a more comprehensive response to environmental changes than is offered by changes in the frequency of already existing alleles.
Evolutionary Adaptation and God’s Providence
From a creation model perspective, the adaptability of organisms is understood as part of the design God ordained in the biological realm. In line with Christian theology, the RTB model maintains that God not only created the world, but he also actively and continually preserves and governs all that he has made. God’s governance of the creation includes the natural processes he instituted when he brought the universe into existence. It is through these processes that he sustains the universe and everything in it.
Evolutionary Adaptation Is Not Evidence for the Evolutionary Paradigm
On the other hand, just because one embraces organisms’ ability to adapt through evolutionary processes, it doesn’t mean they are obligated to accept the totality of the evolutionary paradigm. I don’t.
While abundant evidence exists for microevolution and adaptation (driven by natural and sexual selection, genetic drift, and, now, introgression), it isn’t clear that merely extrapolating these mechanisms over vast time periods can explain large-scale evolutionary change (macroevolution). To put it another way, it isn’t clear if natural and sexual selection, genetic drift, and even adaptive introgression can account for biological novelty and innovation—particularly when life transitions from one regime of complexity to another.
These issues also mean that life scientists cannot legitimately enlist the sound and well-evidenced explanations for microevolution and adaptation in support of macroevolution. They also justify the skepticism that some ID proponents and creationists express about the capacity of evolutionary mechanisms to fully account for the origin, design, and history of life.
I wonder if modern-day biology will be adaptable enough to make a place at the table for ID and creation models—particularly in the face of the shortcomings of current evolutionary theory.
Endnotes
Ming-Shan Wang et al. “A Polar Bear Paleogenome Reveals Extensive Ancient Gene Flow from Polar Bears into Brown Bears,” Nature Ecology and Evolution 6 (June 16, 2022): 936–944, doi:10.1038/s41559-022-01753-8.For example, see Philip W. Hedrick, “Adaptive Introgression in Animals: Examples and Comparison to New Mutation and Standing Variation as a Source of Adaptive Variation,” Molecular Ecology 22, no. 18 (September 2013): 4606–4618, doi:10.1111/mec.12415.News Staff, “Scientists Sequence Genome of 100,000-Year-Old Polar Bear,” Sci News, June 17, 2022, http://www.sci-news.com/genetics/polar-bear-paleogenome-10914.html.Douglas H. Erwin and James W. Valentine, The Cambrian Explosion: The Construction of Animal Biodiversity (Greenwood Village, CO: Roberts and Company, 2013), 10–11.Gerd B. Müller, “Why an Extended Evolutionary Synthesis Is Necessary,” Interface Focus 7 (August 18, 2017): 20170015, doi:10.1098/rsfs.2017.0015.Copyright © 2022 Uncommon Descent . This Feed is for personal non-commercial use only. If you are not reading this material in your news aggregator, the site you are looking at is guilty of copyright infringement UNLESS EXPLICIT PERMISSION OTHERWISE HAS BEEN GIVEN. Please contact legal@uncommondescent.com so we can take legal action immediately.Plugin by Taragana
At Universe Today: Maybe We Don’t See Aliens Because Nobody Wants to Come Here
The Fermi Paradox won’t go away. It’s one of our most compelling thought experiments, and generations of scientists keep wrestling with it. The paradox pits high estimates for the number of civilizations in the galaxy against the fact that we don’t see any of those civs. It says that if rapidly expanding civilizations exist in the Milky Way, one should have arrived here in our Solar System. The fact that none have implies that none exist.
 Artist impression of an alien civilization. Image credit: CfA
Artist impression of an alien civilization. Image credit: CfA
Many thinkers and scientists have addressed the Fermi Paradox and tried to come up with a reason why we don’t see any evidence of an expanding technological civilization. Life may be extraordinarily rare, and the obstacles to interstellar travel may be too challenging. It could be that simple.
But a new paper has a new answer: maybe our Solar System doesn’t offer what long-lived, rapidly expanding civilizations desire: the correct type of star.
To understand the Fermi Paradox, you need to understand the Drake Equation. The Drake Equation is a probabilistic estimate of the number of civilizations in the Milky Way. It doesn’t tell us how many civs there are; it summarizes the concepts we have to wrestle with if we want to think about how many civilizations there could be.
A critical component of the Drake Equation concerns stars. The Equation considers the rate of star formation in the galaxy, how many of those stars host planets, and how many of those planets could host life. The Equation gets more detailed by asking how many of those planets develop life, how much of that life becomes technological civilizations, and how many of those civilizations reveal their presence by releasing signals into space. Finally, it estimates the life spans of those civilizations.
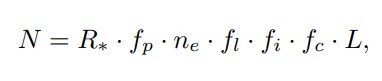 The Drake Equation: Number of Communicative Civilizations (N) = R* (star formation rate) x fp (fraction of stars with planets) x Ne(number of habitable planets per system) x fl (fraction of habitable planets that develop life) x fi (fraction of those that develop intelligent life) x fc (fraction of those that develop communicative technology) x L (average communicative lifetime for those civilizations.)
The Drake Equation: Number of Communicative Civilizations (N) = R* (star formation rate) x fp (fraction of stars with planets) x Ne(number of habitable planets per system) x fl (fraction of habitable planets that develop life) x fi (fraction of those that develop intelligent life) x fc (fraction of those that develop communicative technology) x L (average communicative lifetime for those civilizations.)By using different variables to answer each of those questions, we get different estimates of how many technological civilizations there might be. It’s a thought experiment, but one informed by evidence, though the evidence is rudimentary.
A new paper addresses the Fermi Paradox by focusing on star types. It says that not all types of stars are desirable to an expanding technological civilization. Low-mass stars, particularly K-dwarf stars, are the best migration targets for long-lived civilizations.
The paper is “Galactic settlement of low-mass stars as a resolution to the Fermi paradox,” and the Astrophysical Journal has accepted it for publication. The authors are Jacob Haqq-Misra and Thomas J. Fauchez. Haqq-Misra is a Senior Research Investigator at the Blue Marble Space Institute of Science in Seattle, Washington. Fauchez is a Research Assistant Professor in Physics from the American University in Washington, DC.
The paper begins with a summary of the Fermi Paradox: “An expanding civilization could rapidly spread through the galaxy, so the absence of extraterrestrial settlement in the solar system implies that such expansionist civilizations do not exist,” the authors plainly state.
The authors point to one of the most famous analyses of the Fermi Paradox. It came from American astrophysicist Michael Hart in 1975. Hart’s paper was “An Explanation for the Absence of Extraterrestrials on Earth,” and it was published in the Quarterly Journal of the Royal Astronomical Society. It’s considered to be the first rigorous analysis of the paradox. In his paper, Hart showed how a civilization could expand through the galaxy in a period of time shorter than the galaxy’s age. Hart explained what would happen if a civilization sent out colony ships to the nearest 100 stars. They could colonize those star systems, then each of those colonies could do the same, and the process could keep repeating.
Copyright © 2022 Uncommon Descent . This Feed is for personal non-commercial use only. If you are not reading this material in your news aggregator, the site you are looking at is guilty of copyright infringement UNLESS EXPLICIT PERMISSION OTHERWISE HAS BEEN GIVEN. Please contact legal@uncommondescent.com so we can take legal action immediately.“If there were no pause between trips, the frontier of space exploration would then lie roughly on the surface of a sphere whose radius was increasing at a speed of 0.10c,” Hart wrote. “At that rate, most of our Galaxy would be traversed within 650,000 years.” Hart pointed out that a technological civilization would’ve had ample time to reach us unless they had started less than two million years ago. For Hart, the only explanation for the lack of evidence of alien civilizations is that there are none.
In his paper, Hart arrived at a couple of conclusions: SETI and similar efforts are a waste of time and money, and if anyone colonizes our Solar System, it’ll probably be our descendants who do it.
See the rest of the article at Universe Today.
Plugin by Taragana
October 27, 2022
From The Wall Street Journal: ‘Imperfection’ Review: Unintelligent Design
David P. Barash writes:
‘Imperfection,” by Telmo Pievani, begins as it should, with the big bang: “In the beginning, there was imperfection. A rebellion against the established order, with no witnesses, in the heart of the darkest of nights. Something in the symmetry broke down 13.82 billion years ago.” And it ends on a suitably ambiguous note: “There is something amazing in evolution . . . which in 3.5 billion years has taken us from an amoeba to Donald Trump.”
 Credit: PEERAGRIT, 123rf.com
Credit: PEERAGRIT, 123rf.comMr. Pievani is a professor of biology at the University of Padua. His brief and thoughtful book (translated from the Italian by Michael Gerard Kenyon) isn’t just a description of imperfection, but a paean to it. There’s plenty of description and discussion, too, as “Imperfection” takes the reader on a convincing whirlwind tour of the dangers as well as the impossibility of perfection, how imperfection is built into the nature of the universe, and into all living things—including ourselves.
Mary Poppins congratulated herself for being “practically perfect in every way,” but of course she wasn’t, if only because she bragged about it. Moreover, perfection would make evolution stop dead in its tracks. In fact, it would never have begun—natural selection needs diversity upon which to operate. And diversity ultimately arises from mutation and sexual recombination, each of which is a perfect source of imperfection.
Yet another source of imperfection, unique to Homo sapiens and well described by Mr. Pievani, is the disconnect between our rapid cultural innovation and our slow biological evolution. (Immodest note: In 1986, your current reviewer wrote “The Hare and the Tortoise,” the first book calling attention to this troublesome imbalance.) For a homey example, consider that, being primates, our Pleistocene ancestors were naturally fond of sugars, which indicate ripe fruit, and of fats, present—albeit in generally small quantities—in game. Today, our culture provides us with excessive opportunities to indulge such fondness, which we overdo, benefiting only the confectionery and meat industries, along with dentists, cardiologists and morticians.
Abraham Lincoln had a cute way of undercutting our tendency to find perfection everywhere. It’s remarkable, he once pointed out, that no matter how tall someone is, their legs are always exactly long enough to reach the ground! Ironically, fundamentalists on both extremes of the evolution divide often converge in misinterpreting perfection, creationists proclaiming that only a supreme being could have produced such superb complexity, while hyper-adaptationists emphasize the power of natural selection to achieve the same thing, promoting a “gee whiz” perspective on evolution.
Counterintuitively, it is the imperfection of the organic world that provides some of the most cogent evidence for evolution as a wholly natural phenomenon, and against special creation, or, in its barely disguised incarnation,“intelligent design theory.” And here is where “Imperfection,” the book, is especially valuable.
As Mr. Pievani emphasizes, Homo sapiens are marvels of unintelligent design, “with their useless earlobes, their tedious wisdom teeth, . . . their vermiform intestinal appendage, their spinal curves, and their vas deferens, which carries sperm from the testicles to the penis not directly and by the shortest route but instead after going by a useless and lengthy route via the ureter . . . the remains of their ancestral quadrupedal gait, and the corresponding ills and pains, backache, sciatica, flat feet, scoliosis, and hernias.” Add the terrible structure of our knees, our lower backs, the fact that the opening of the tubes carrying food and air are so close that choking is a significant cause of mortality, the awkwardness of having our reproduction and sewerage emerging right next to each other. We are shot through with deficiencies that wouldn’t earn even a passing grade for a novice bioengineer, never mind an omniscient, omnipotent deity.
Full article at The Wall Street Journal.
So, this article continues, but let’s not overlook the grandeur of a mountain by having one’s eyes focused downward on a crack in a rock. Anyone who says, “Homo sapiens are marvels of unintelligent design”, and can only myopically see subjectively critiqued imperfections, while not being awed by the suite of biochemical marvels that comprise the human body, is ignorant at best, and perhaps maliciously antagonistic towards God at worst.
Copyright © 2022 Uncommon Descent . This Feed is for personal non-commercial use only. If you are not reading this material in your news aggregator, the site you are looking at is guilty of copyright infringement UNLESS EXPLICIT PERMISSION OTHERWISE HAS BEEN GIVEN. Please contact legal@uncommondescent.com so we can take legal action immediately.Plugin by Taragana
At Phys.org: Aluminous silica: A major water carrier in the lower mantle
Water is transported by oceanic plates into the Earth’s deep interior and changes the properties of minerals and rocks, affecting the Earth’s internal material cycle and environmental evolution since the formation of the Earth.
 Mantle convection including water by hydrous aluminous silicas. Credit: Takayuki Ishii
Mantle convection including water by hydrous aluminous silicas. Credit: Takayuki IshiiAn international research group…reveals that aluminous silicas play a major role as a water carrier in the lower mantle. They determined the alumina and water contents of silica minerals, which are important minerals in the basaltic crust of the upper part of a subducting plate.
The results show that rutile-type silica (stishovite), which is widely stable in the upper part of the lower mantle, undergoes a phase transition to a CaCl2-type phase, when it contains water and alumina. The CaCl2-type aluminous silica can hold more than 10 times the amount of water than other lower mantle minerals, even at very high temperatures in the lower mantle.
Since the birth of the Earth, water has traveled through the Earth’s surface and interior, triggering earthquakes and volcanic activity and affecting the evolution of the Earth’s interior environment. It is estimated that the amount of water that can be stored in the Earth’s interior is several times that of the seawater on the Earth’s surface.
Water (seawater) is transported to the Earth’s interior by oceanic plates. To prevent water from leaking out of the plates, the minerals that make up the plates efficiently transport water by incorporating it into their crystal structures. It is thought that the minerals are transported by the plates to the lower mantle and then returned to the Earth’s surface by the upwelling plume. It is still not well understood how much water is stored in the Earth’s interior and how it returns to the Earth’s surface. To understand these issues, it is important to know how much water mantle minerals can contain and how stably they can hold water.
Previously reported mantle minerals release water with increasing temperature: their water content generally decreases with temperature. Since temperature increases with depth in the Earth’s interior, this property implies that the water-holding capacity of minerals decreases with depth.
When water is released from minerals, it reacts with rocks to form hydrous magma, which separates from the plate and moves to the surface. Therefore, the depth at which minerals release water is considered to be the upper limit of water transport depth. It has been pointed out that in the lower mantle, which is particularly hot, minerals cannot hold water and may not be able to transport water. Contrary to this property, the minerals synthesized in this study have a water content that increases with temperature and can hold large amounts of water even under the hottest plume conditions in the mantle.
Full article at Phys.org.
Earth’s “deep water cycle,” which transports water from the crust and ocean into the mantle and back, has been recognized as a key feature in supporting optimum plate tectonics to sustain Earth’s habitability. See Reasons.org for an additional article on this topic.
Copyright © 2022 Uncommon Descent . This Feed is for personal non-commercial use only. If you are not reading this material in your news aggregator, the site you are looking at is guilty of copyright infringement UNLESS EXPLICIT PERMISSION OTHERWISE HAS BEEN GIVEN. Please contact legal@uncommondescent.com so we can take legal action immediately.Plugin by Taragana
Peto’s paradox – how intelligent design solves it
Peto’s paradox – how intelligent design solves it
Marc Tollis (2017): In a multicellular organism, cells must go through a cell cycle that includes growth and division. Every time a human cell divides, it must copy its six billion base pairs of DNA, and it inevitably makes some mistakes. These mistakes are called somatic mutations (cells in the body other than sperm and egg cells). Some somatic mutations may occur in genetic pathways that control cell proliferation, DNA repair, apoptosis, telomere erosion, and growth of new blood vessels, disrupting the normal checks on carcinogenesis. If every cell division carries a certain chance that a cancer-causing somatic mutation could occur, then the risk of developing cancer should be a function of the number of cell divisions in an organism’s lifetime. Therefore, large-bodied and long-lived organisms should face a higher lifetime risk of cancer simply due to the fact that their bodies contain more cells and will undergo more cell divisions over the course of their lifespan. However, a 2015 study that compared cancer incidence from zoo necropsy data for 36 mammals found that a higher risk of cancer does not correlate with increased body mass or lifespan. In fact, the evidence suggested that larger long-lived mammals actually get less cancer. This has profound implications for our understanding of how the cancer problem is solved.
When individuals in populations are exposed to the selective pressure of cancer risk, the population must evolve cancer suppression as an adaptation or else suffer fitness costs and possibly extinction. Discovering the mechanisms underlying these solutions to Peto’s Paradox requires the tools of numerous subfields of biology including genomics, comparative methods, and experiments with cells. For instance, genomic analyses revealed that the African savannah elephant (Loxodonta africana) genome contains 20 copies, or 40 alleles, of the most famous tumor suppressor gene TP53. The human genome contains only one TP53 copy, and two functional TP53 alleles are required for proper checks on cancer progression. When cells become stressed and incur DNA damage, they can either try to repair the DNA or they can undergo apopotosis, or self-destruction. The protein produced by the TP53 gene is necessary to turn on this apoptotic pathway. Humans with one defective TP53 allele have Li Fraumeni syndrome and a ~90% lifetime risk of many cancers, because they cannot properly shut down cells with DNA damage. Meanwhile, experiments revealed that elephant cells exposed to ionizing radiation behave in a manner consistent with what you would expect with all those TP53 copies—they are much more likely to switch on the apoptotic pathway and therefore destroy cells rather than accumulate carcinogenic mutations. 1
Comment: How does the author explain the origin of these protective mechanisms? He claims: “The solution to Peto’s Paradox is quite simple: evolution”. This is an ad-hoc assertion and raises the question: How could complex multicellular organisms have evolved if these cancer protection mechanisms were not implemented before the transition occurred, since, otherwise, these organisms would have gone extinct? This problem becomes even greater if considering, that animals with large body size supposedly evolved independently many times across the history of life, and, therefore, these mechanisms would have had to be recruited multiple times. The paradox is only solved, if we hypothesize that large animals were created independently by God, and right from the beginning equipped with tumor suppressor mechanisms from the get-go.
A. B. Williams (2016): The loss of p53 is a major driver of cancer development mainly because, in the absence of this “guardian of the genome,” cells are no longer adequately protected from mutations and genomic aberrations. Intriguingly, the evolutionary occurrence of p53 homologs appears to be associated with multicellularity. With the advent of metazoans, genome maintenance became a specialized task with distinct requirements in germ cells and somatic tissues. With the central importance of p53 in controlling genome instability–driven cancer development, it might not be surprising that p53 controls DNA-damage checkpoints and impacts the activity of various DNA-repair systems. 2
B. J. Aubrey (2016): The fundamental biological importance of the Tp53 gene family is highlighted by its evolutionary conservation for more than one billion years dating back to the earliest multicellular organisms. The TP53 protein provides essential functions in the cellular response to diverse stresses and safeguards maintenance of genomic integrity, and this is manifest in its critical role in tumor suppression. The importance of Tp53 in tumor prevention is exemplified in human cancer where it is the most frequently detected genetic alteration. This is confirmed in animal models, in which a defective Tp53 gene leads inexorably to cancer development, whereas reinstatement of TP53 function results in regression of established tumors that had been initiated by loss of TP53.
TP53: Tumor Suppression and Transcriptional Regulation
Following activation, the TP53 protein functions predominantly as a transcription factor. The TP53 protein forms a homotetramer that binds to specific Tp53 response elements in genomic DNA to direct the transcription of a large number of protein-coding genes. The requirement for TP53 transcriptional activity in tumor suppression has been examined by systematically mutating the transactivation domains of the TP53 protein, rendering it either partially or wholly transcriptionally defective. Importantly, mutations resulting in complete loss of TP53 transcriptional activity ablate its ability to prevent tumor formation, supporting the concept that transcriptional regulation is central to the tumor-suppressor function. TP53-mediated tumor suppression is governed by transcriptional regulation.
TP53-mediated transcriptional regulation varies according to the type of stress stimulus and type of cell, so that appropriate corrective processes can be implemented. For example, minor DNA damage may institute cell-cycle arrest and activate DNA-repair mechanisms, whereas stronger TP53-activating signals induce senescence or apoptosis. Accordingly, the TP53 transcriptional response varies depending on the nature of the activating signal and the type of cell. The number of known or suspected TP53 target genes has increased into the thousands with dramatic differences in transcriptional responses observed among different cell types, different TP53-inducing stress stimuli, and varying time points following TP53 activation. These studies paint an increasingly complex picture of the modes by which TP53 can regulate gene expression. For example, before TP53 activation, a subset of target genes is transcriptionally repressed by the TP53 protein. More recently appreciated functions of the TP53 protein include widespread binding and modulation of enhancer regions throughout the genome and transcriptional activation of noncoding RNAs. Interestingly, the TP53-activated long noncoding RNA, lincRNA-p21, exerts widespread suppression of gene expression. The list of proposed TP53 target genes is vast and they are known to influence diverse cellular processes, including apoptosis, cell-cycle arrest, senescence, DNA-damage repair, metabolism, and global regulation of gene expression, each of which could potentially contribute to its tumor-suppressor function.
The p53 pathway responds to various cellular stress signals (the input) by activating p53 as a transcription factor (increasing its levels and protein modifications) and transcribing a programme of genes (the output) to accomplish a number of functions. Together, these functions prevent errors in the duplication process of a cell that is under stress, and as such the p53 pathway increases the fidelity of cell division and prevents cancers from arising. 3
K. D. Sullivan (2017): The p53 polypeptide contains several functional domains that work coordinately, in a context-dependent fashion, to achieve DNA binding and transactivation. (the increased rate of gene expression) 4
K. Kamagata (2020): Interactions between DNA and DNA-binding proteins play an important role in many essential cellular processes. A key function of the DNA-binding protein p53 is to search for and bind to target sites incorporated in genomic DNA, which triggers transcriptional regulation. How do p53 molecules achieve “rapid” and “accurate” target search in living cells? The genome encompasses DNA sequences that encode genes, and gene editing is the genetic engineering of a specific DNA sequence, including insertion, deletion, modification, and replacement. The main player in genome editing is a type of protein that can bind to DNA, known as DNA-binding proteins. DNA-binding proteins include enzymes, which can cut DNA or ligate two DNA molecules, and transcription factors, which can activate or deactivate gene expression. These proteins are classified into DNA sequence-specific and nonspecific binders. The transcription factor p53 can induce multiple tumor suppression functions, such as cell cycle arrest, DNA repair, and apoptosis. p53 is presumed to solve the target search problem by utilizing 3D diffusion, 1D diffusion along DNA, and intersegmental transfer between two DNAs in the cell. 5
Comment: This transcription factor p53 actively searches targets in the genome to be expressed. This is a goal-oriented process implemented to activate processes that avoid the origination of cancer. Various players are required that work as a system. It is a team play. The p53 transcription factor has to be able to perform “rapid” and “accurate” target search, recognize it, and bind to the DNA sequence so it can be expressed, but most important, before it can act like a switch commanding “on”, the gene sequences to be expressed must be there, that is, the actors that are recruited to permit DNA repair, or apoptosis (cell death). It is an all-or-nothing business to convey the function to suppress the development and growth of tumors, and consequently, death. In other words, this is an irreducibly complex system where p53 would be functionless unless the actors to act upon were not there.
The concepts of machine and factory error monitoring, checking, and repair are all tasks performed with goal-directedness, intent, and purpose.
1. Repairing things that are broken, malfunctioning, or instantiating complex systems that autonomously prevent things to break are always actions performed by agents with intentions, volition, goal-orientedness, foresight, understanding, and know-how.
2. Man-made machines almost always require direct intelligent intervention by technicians to recognize errors, find which parts of a machine are broken, know how to remove and replace them without breaking surrounding parts of the device, and know how to construct the part that has to be replaced with fidelity, and re-insert and re-connect it where the part was removed. The entire process is complex, demanding know-how, and depends on a high quantity of intelligence in performing all involved actions.
3. Man has not been able to create a fully autonomous, preprogrammed machine or factory, that is able to quality and error monitor all manufacturing processes and the correct performance of all devices involved, and if the products are up to the required quality standard, and, if something drives havoc, repair and re-establish normal function of what was broken or malfunctioning without external intervention.
4. C.H. Loch writes in the science paper: “Organic Production Systems: What the Biological Cell Can Teach Us About Manufacturing” (2004): Biological cells are preprogrammed to use quality-management techniques used in manufacturing today. The cell invests in defect prevention at various stages of its replication process, using 100% inspection processes, quality assurance procedures, and foolproofing techniques. An example of the cell inspecting each and every part of a product is DNA proofreading. As the DNA gets replicated, the enzyme DNA polymerase adds new nucleotides to the growing DNA strand, limiting the number of errors by removing incorrectly incorporated nucleotides with a proofreading function. Following is an impressive example: Unbroken DNA conducts electricity, while an error blocks the current. Some repair enzymes exploit this. One pair of enzymes lock onto different parts of a DNA strand. One of them sends an electron down the strand. If the DNA is unbroken, the electron reaches the other enzyme and causes it to detach. I.e. this process scans the region of DNA between them, and if it’s clean, there is no need for repairs. But if there is a break, the electron doesn’t reach the second enzyme. This enzyme then moves along the strand until it reaches the error, and fixes it. This mechanism of repair seems to be present in all living things, from bacteria to man. Know-how is needed:
Martin_r:
a. To know that something is broken (DNA damage sensing)
b. To identify where exactly it is broken
c. To know when to repair it (e.g. one has to stop/or put on hold some other ongoing processes, in other words, one needs to know lots of other things, one needs to know the whole system, otherwise one creates more damage…)
d. to know how to repair it (to use the right tools, materials, energy, etc, etc, etc )
e. to make sure that the repair was performed correctly. (this can be observed in DNA repair as well)
5. On top of that: Cells do not even wait until a protein machine fails, but replace it long before it has a chance to break down. Furthermore, it completely recycles the machine that is taken out of production. The components derived from this recycling process can be used not only to create other machines of the same type but also to create different machines if that is what is needed in the “plant.” This way of handling its machines has some clear advantages for the cell. New capacity can be installed quickly to meet current demand. At the same time, there are never idle machines around taking up space or hogging important building blocks. Maintenance is a positive “side effect” of the continuous machine renewal process, thereby guaranteeing the quality of output. Finally, the ability to quickly build new production lines from scratch has allowed the cell to take advantage of a big library of contingency plans in its DNA that allow it to quickly react to a wide range of circumstances, as we will describe next.
6. The more sophisticated, advanced, autonomous, complex, and information-driven machines or factories are, the more they carry the hallmark of design. The very concepts of error monitoring, checking, and repair, and replacement in advance to avoid future break-ups are tasks performed with goal-directedness, and purpose. Biological cells are far more advanced than any machine and factory ever devised and invented by man. It is therefore rational and warranted to infer, that biological cells were designed.
Plugin by Taragana
October 26, 2022
At Evolution News: The Standard Story of Human Evolution: A Critical Look
Casey Luskin writes:
Despite disagreements, there is a standard story of human evolution that is retold in countless textbooks, news media articles, and documentaries. Indeed, virtually all the scientists I am citing here accept some evolutionary account of human origins, albeit flawed.
Starting with the early hominins and moving through the australopithecines, and then into the genus Homo, I will review the fossil evidence and assess whether it supports this standard account of human evolution. As we shall see, the evidence — or lack thereof — often contradicts this evolutionary story.
 Photo: Ardipithecus ramidus, by Tiia Monto, CC BY-SA 3.0 , via Wikimedia Commons.Early Hominins
Photo: Ardipithecus ramidus, by Tiia Monto, CC BY-SA 3.0 , via Wikimedia Commons.Early HomininsIn 2015, two leading paleoanthropologists reviewed the fossil evidence regarding human evolution in a prestigious scientific volume titled Macroevolution. They acknowledged the “dearth of unambiguous evidence for ancestor-descendant lineages,” and admitted,
[T]he evolutionary sequence for the majority of hominin lineages is unknown. Most hominin taxa, particularly early hominins, have no obvious ancestors, and in most cases ancestor-descendant sequences (fossil time series) cannot be reliably constructed.1
Nevertheless, numerous theories have been promoted about early hominins and their ancestral relationships to humans.
One leading fossil is described below:
Ardipithecus ramidus: Irish Stew or Breakthrough of the Year?
In 2009, Science announced the long-awaited publication of details about Ardipithecus ramidus (pictured above), a would-be hominin fossil that lived about 4.4 million years ago (mya). Expectations mounted after its discoverer, UC Berkeley paleoanthropologist Tim White, promised a “phenomenal individual” that would be the “Rosetta stone for understanding bipedalism.”17 The media eagerly employed the hominin they affectionately dubbed Ardi to evangelize the public for Darwin.
Discovery Channel ran the headline “‘Ardi,’ Oldest Human Ancestor, Unveiled,” and quoted White calling Ardi “as close as we have ever come to finding the last common ancestor of chimpanzees and humans.”18 The Associated Press declared, “World’s Oldest Human-Linked Skeleton Found,” and stated that “the new find provides evidence that chimps and humans evolved from some long-ago common ancestor.”19 Science named Ardi the “breakthrough of the year” for 2009,20 and introduced her with the headline, “A New Kind of Ancestor: Ardipithecus Unveiled.”21
Calling Ardi “new” may have been a poor word choice, for it was discovered in the early 1990s. Why did it take some 15 years to publish the analyses? A 2002 article in Science explains the bones were “soft,” “crushed,” “squished,” and “chalky.”22 Later reports similarly acknowledged that “portions of Ardi’s skeleton were found crushed nearly to smithereens and needed extensive digital reconstruction,” including the pelvis, which “looked like an Irish stew.”23
Claims about bipedal locomotion require accurate measurements of the precise shapes of key bones (like the pelvis). Can one trust declarations of a “Rosetta stone for understanding bipedalism” when Ardi was “crushed to smithereens”? Science quoted various paleoanthropologists who were “skeptical that the crushed pelvis really shows the anatomical details needed to demonstrate bipedality.”24
Even some who accepted Ardi’s reconstructions weren’t satisfied that the fossil was a bipedal human ancestor. Primatologist Esteban Sarmiento concluded in Science that “[a]ll of the Ar. ramidus bipedal characters cited also serve the mechanical requisites of quadrupedality, and in the case of Ar. ramidus foot-segment proportions, find their closest functional analog to those of gorillas, a terrestrial or semiterrestrial quadruped and not a facultative or habitual biped.”25 Bernard Wood questioned whether Ardi’s postcranial skeleton qualified it as a hominin,26 and co-wrote in Nature that if “Ardipithecus is assumed to be a hominin,” then it had “remarkably high levels of homoplasy [similarity] among extant great apes.”27 A 2021 study found that Ardi’s hands were well-suited for climbing and swinging in trees, and for knuckle-walking, giving it a chimp-like mode of locomotion.28 In other words, Ardi had ape-like characteristics which, if we set aside the preferences of Ardi’s promoters, should imply a closer relationship to apes than to humans. As the authors of the Nature article stated, Ardi’s “being a human ancestor is by no means the simplest, or most parsimonious explanation.”29Sarmiento even observed that Ardi had characteristics different from both humans and African apes, such as its unfused jaw joint, which ought to remove her far from human ancestry.30
Whatever Ardi was, everyone agrees the fossils was initially badly crushed and needed extensive reconstruction. No doubt this debate will continue, but are we obligated to accept the “human ancestor” position promoted by Ardi’s discoverers in the media? Sarmiento doesn’t think so. According Time magazine, he “regards the hype around Ardi to have been overblown.”31
Full article at Evolution News.
NotesBernard Wood and Mark Grabowski, “Macroevolution in and around the Hominin Clade,” Macroevolution: Explanation, Interpretation, and Evidence, eds. Serrelli Emanuele and Nathalie Gontier (Heidelberg, Germany: Springer, 2015), 347-376.Michel Brunet et al., “Sahelanthropus or ‘Sahelpithecus’?,” Nature 419 (October 10, 2002), 582.Michel Brunet et al., “A new hominid from the Upper Miocene of Chad, Central Africa,” Nature 418 (July 11, 2002), 145-151. See also Michel Brunet et al., “New material of the earliest hominid from the Upper Miocene of Chad,” Nature 434 (April 7, 2005), 752-755. Smithsonian Natural Museum of Natural History, “Sahelanthropus tchadensis,” https://humanorigins.si.edu/evidence/... (accessed November 30, 2020).“Skull Find Sparks Controversy,” BBC News (July 12, 2002), http://news.bbc.co.uk/2/hi/science/na... (accessed October 26, 2020).Milford Wolpoff et al., “Sahelanthropus or ‘Sahelpithecus’?” Nature 419 (October 10, 2002), 581-582.Roberto Macchiarelli et al., “Nature and relationships of Sahelanthropus tchadensis,” Journal of Human Evolution 149 (2020), 102898.Macchiarelli et al., “Nature and relationships of Sahelanthropus tchadensis.”Madelaine Böhme, quoted in Michael Marshall, “Our supposed earliest human relative may have walked on four legs,” New Scientist (November 18, 2020), https://www.newscientist.com/article/... (accessed November 30, 2020).Bob Yirka, “Study of partial left femur suggests Sahelanthropus tchadensis was not a hominin after all,” Phys.org (November 24, 2020), https://phys.org/news/2020-11-partial... (accessed November 30, 2020).Potts and Sloan, What Does It Mean to Be Human?, 38.John Noble Wilford, “Fossils May Be Earliest Human Link,” New York Times (July 12, 2001), http://www.nytimes.com/2001/07/12/wor... (accessed October 26, 2020).John Noble Wilford, “On the Trail of a Few More Ancestors,” New York Times (April 8, 2001), http://www.nytimes.com/2001/04/08/wor... (accessed October 26, 2020).Leslie Aiello and Mark Collard, “Our Newest Oldest Ancestor?” Nature 410 (March 29, 2001), 526-527.K. Galik et al., “External and Internal Morphology of the BAR 1002’00 Orrorin tugenensis Femur,” Science 305 (September 3, 2004), 1450-1453.Sarmiento, Sawyer, and Milner, The Last Human, 35.Tim White, quoted in Ann Gibbons, “In Search of the First Hominids,” Science 295 (February 15, 2002), 1214-1219.Jennifer Viegas, “‘Ardi,’ Oldest Human Ancestor, Unveiled,” Discovery News (October 1, 2009), https://web.archive.org/web/201106130... (accessed October 26, 2020).Randolph Schmid, “World’s Oldest Human-Linked Skeleton Found,” NBC News (October 1, 2009), https://www.nbcnews.com/id/wbna33110809 (accessed October 26, 2020). Ann Gibbons, “Breakthrough of the Year: Ardipithecus ramidus,” Science 326 (December 18, 2009), 1598-1599.Gibbons, “New Kind of Ancestor,” 36-40.White, quoted in Gibbons, “In Search of the First Hominids,” 1214-1219, 1215-1216.Michael Lemonick and Andrea Dorfman, “Ardi Is a New Piece for the Evolution Puzzle,” Time (October 1, 2009), http://content.time.com/time/magazine... (accessed October 26, 2020).Gibbons, “New Kind of Ancestor,” 36-40, 39.Esteban Sarmiento, “Comment on the Paleobiology and Classification of Ardipithecus ramidus,” Science 328 (May 28, 2010), 1105b.Gibbons, “New Kind of Ancestor,” 36-40.Bernard Wood and Terry Harrison, “The Evolutionary Context of the First Hominins,” Nature 470 (February 17, 2011), 347-352.Thomas C. Prang, Kristen Ramirez, Mark Grabowski, and Scott A. Williams, “Ardipithecus hand provides evidence that humans and chimpanzees evolved from an ancestor with suspensory adaptations,” Science Advances 7 (February 24, 2021), eabf2474.New York University, “Fossils may look like human bones: Biological anthropologists question claims for human ancestry,” ScienceDaily (February 16, 2011), https://www.sciencedaily.com/releases... (accessed October 26, 2020).See Eben Harrell, “Ardi: The Human Ancestor Who Wasn’t?,” Time (May 27, 2010), http://content.time.com/time/health/a... (accessed October 26, 2020).Harrell, “Ardi: The Human Ancestor Who Wasn’t?”Copyright © 2022 Uncommon Descent . This Feed is for personal non-commercial use only. If you are not reading this material in your news aggregator, the site you are looking at is guilty of copyright infringement UNLESS EXPLICIT PERMISSION OTHERWISE HAS BEEN GIVEN. Please contact legal@uncommondescent.com so we can take legal action immediately.Plugin by Taragana
Michael J. Behe's Blog
- Michael J. Behe's profile
- 219 followers



